A Little About Me
Welcome to my personal website !
I am a 4th-year cs phd student at Kim’s lab at the University of Toronto, specializing in AI for science. My research focuses on using generative AI to design pharmaceutical peptide and protein drugs, with projects including macrocyclic peptides drugs, D-peptide drugs and epitope-specific antibodies for applications such as wet-lab validated GLP-1 agonists and SARS-CoV-2 treatments.
My research focuses on therapeutic peptides and antibody design, where I integrate computational and experimental methods to address pressing biomedical challenges. Over the past four years, I’ve developed and applied deep learning models, including flow-matching, score-based diffusion models, GANs, and transformers, to design and validate novel therapeutics.
Check out my publication page for more details.
Talks & Research Updates
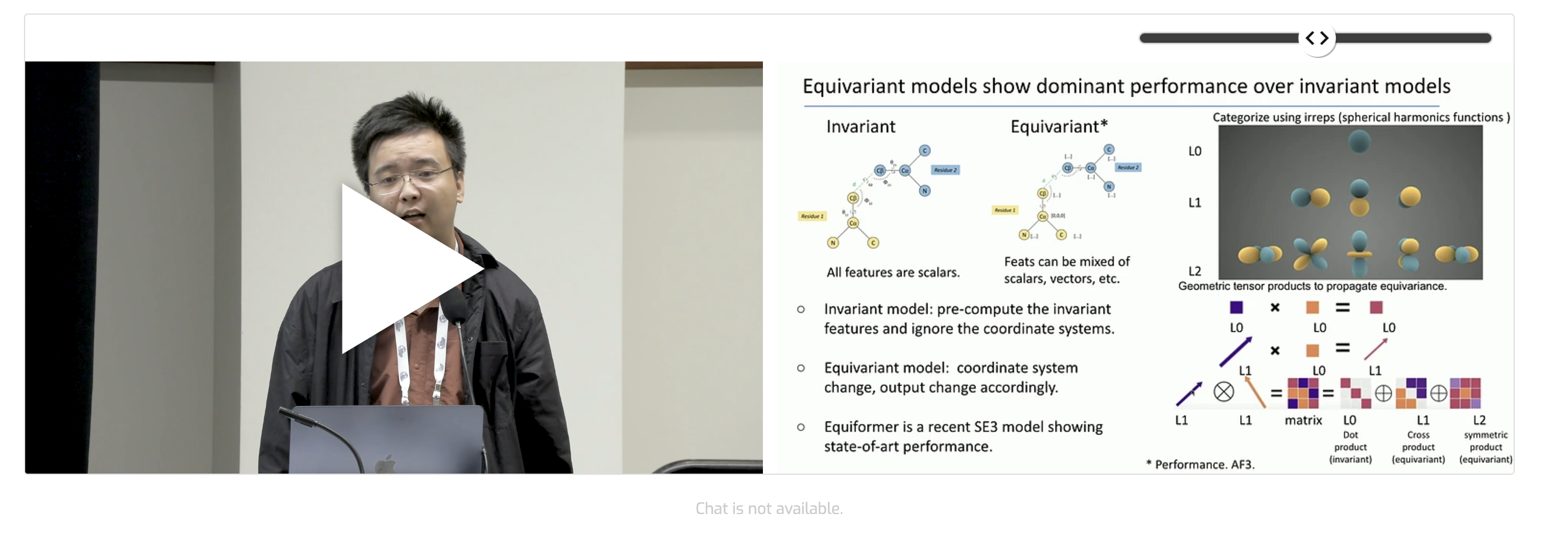
I’m honored to be selected for an oral presentation at MLSB NeurIPS 2024 last December, where I discussed peptide design using an E3NN Flow-Matching model. Check out the link below for details on the talk!
Check out my new paper using diffusion model for peptide drug design as well as our designed Art Work Which Was Selected as Cover Figure for ACS Science (IF 18.2). On the right side, there was my Recorded Virtual Talk on MLSB NeurIPS regarding peptide design using generative model (HelixGAN)
Check out my recent work on Flow-matching model using E3NN to direct monitering atom movements. Flow Matching (FM) is a recent generative modeling approach gaining popularity in the ML community. It combines elements from flow models and diffusion models (DMs), addressing their drawbacks. The Gif for Flow-matching Process inide HelixFlow Model (14, 16, 18, 20 AA)
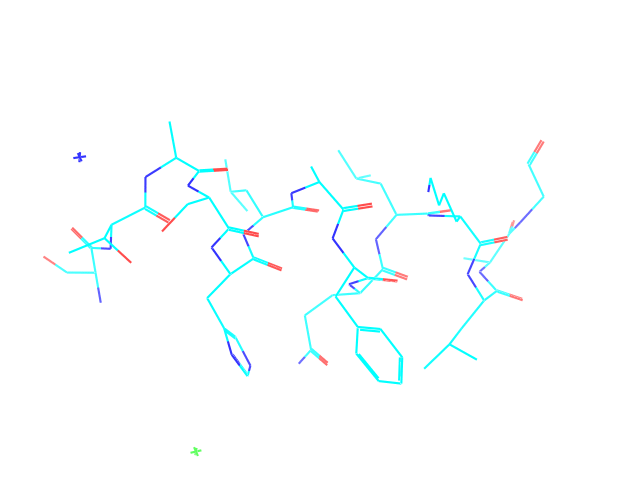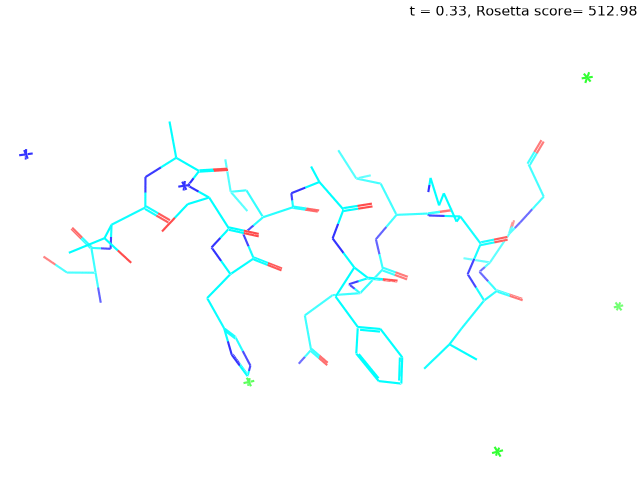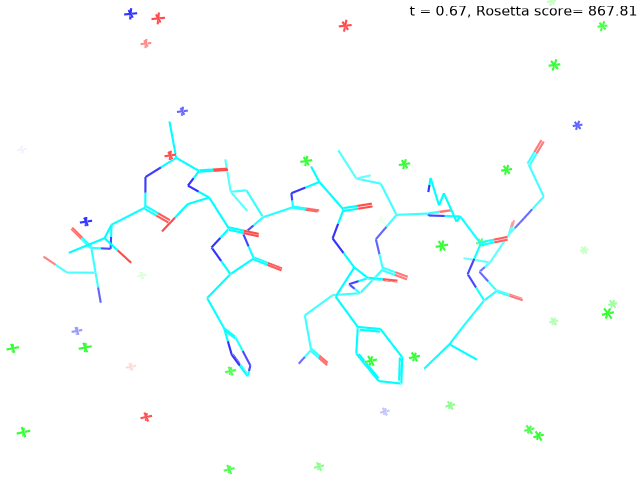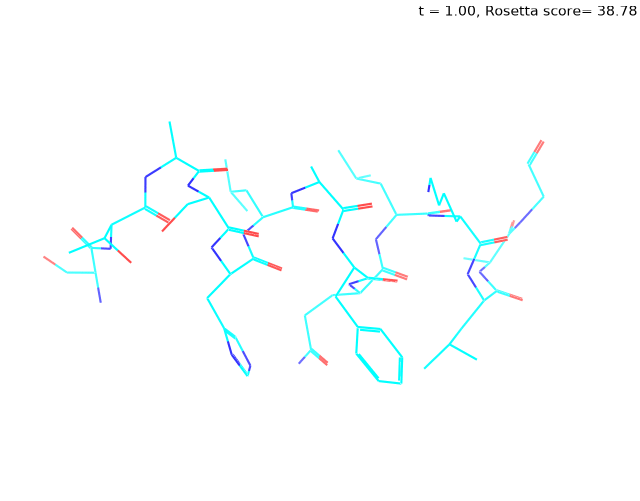
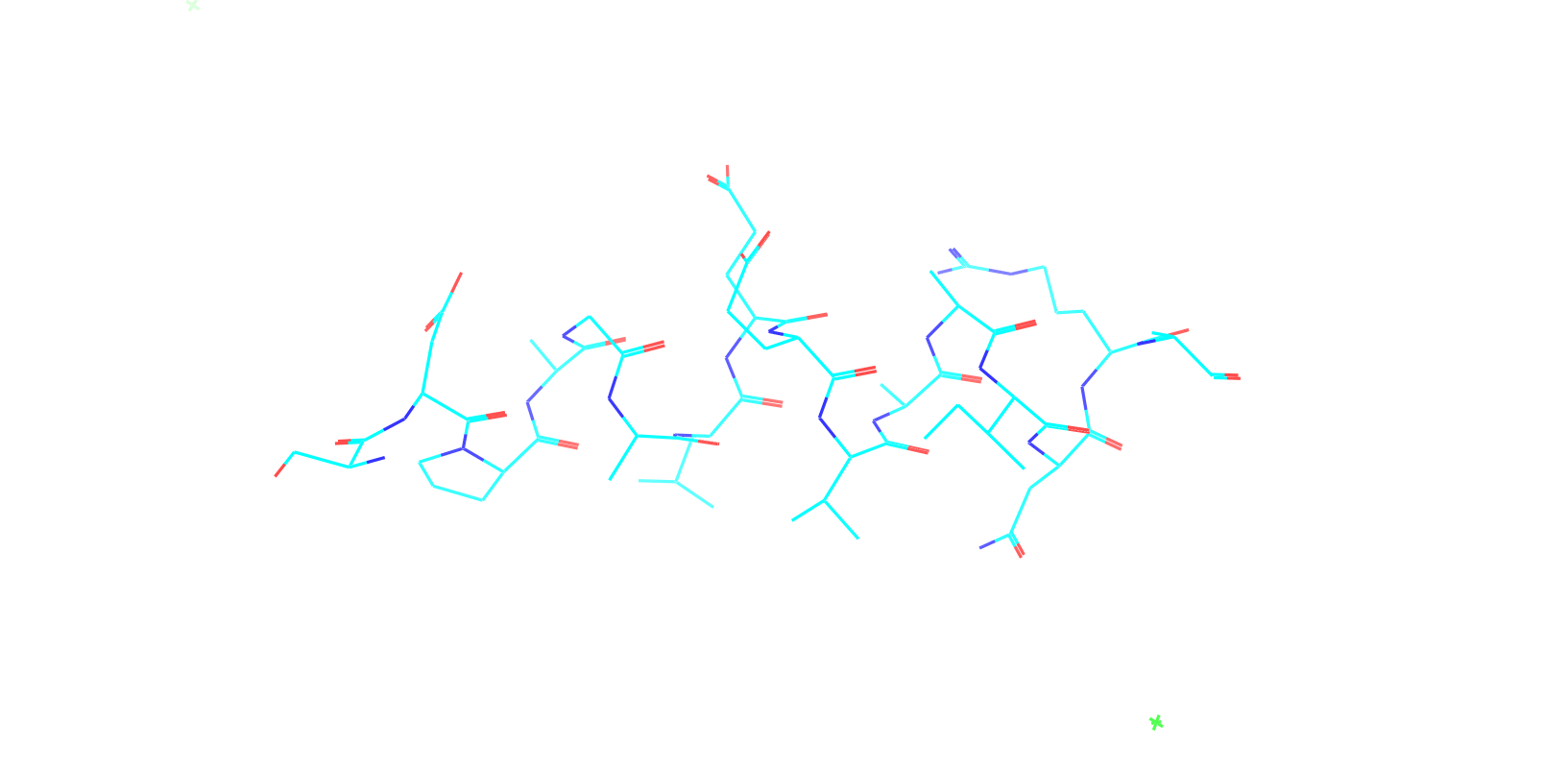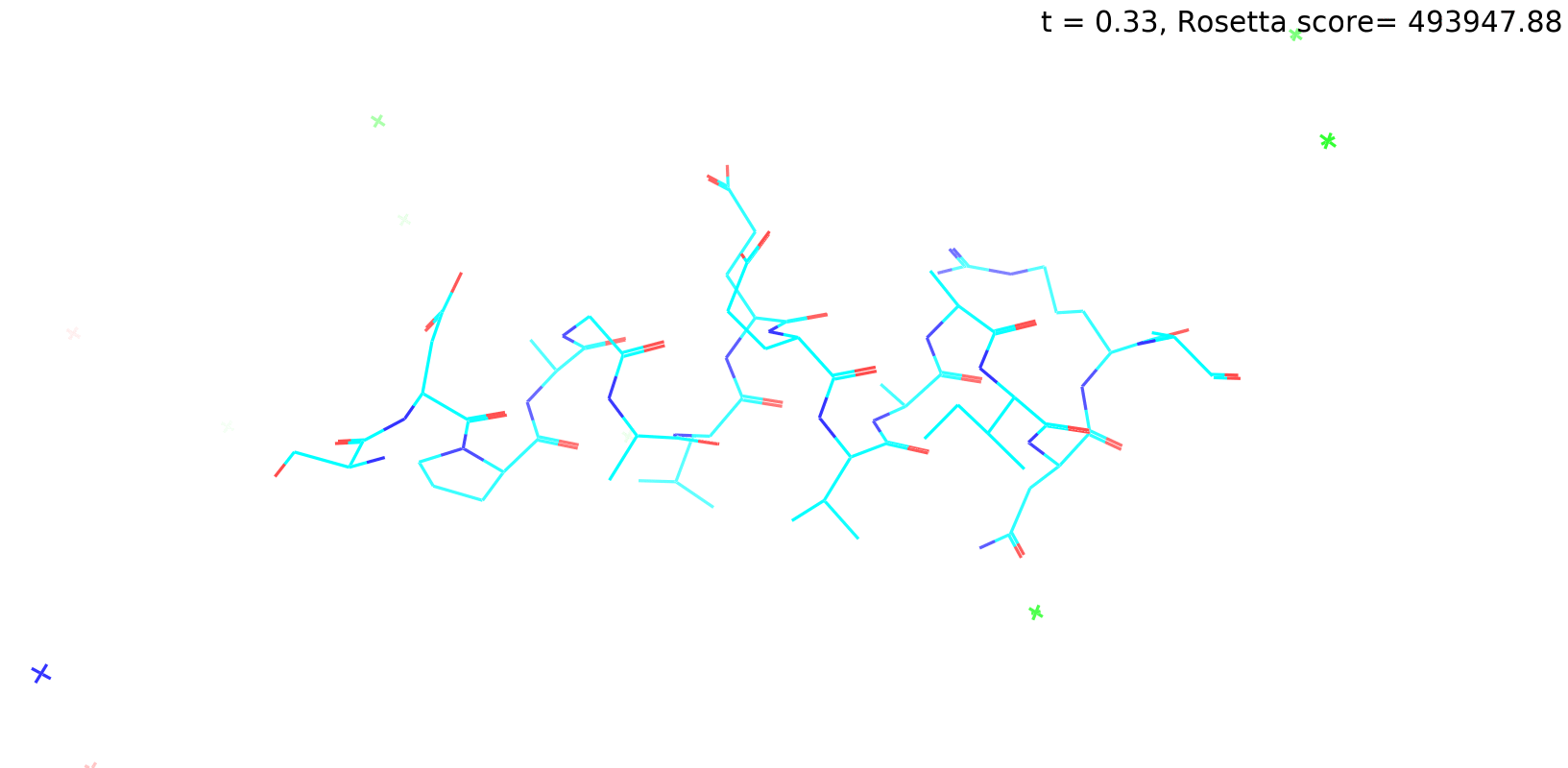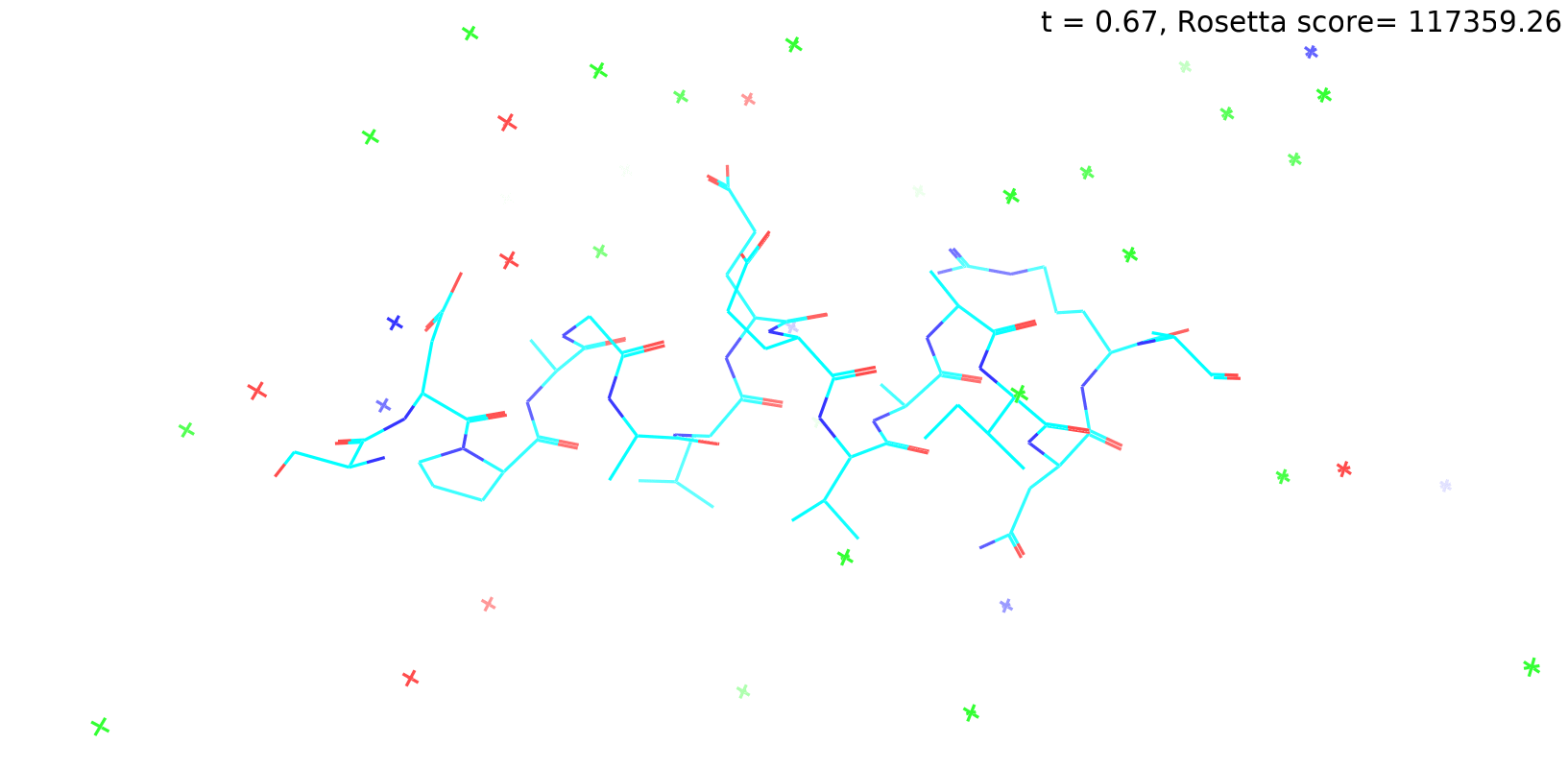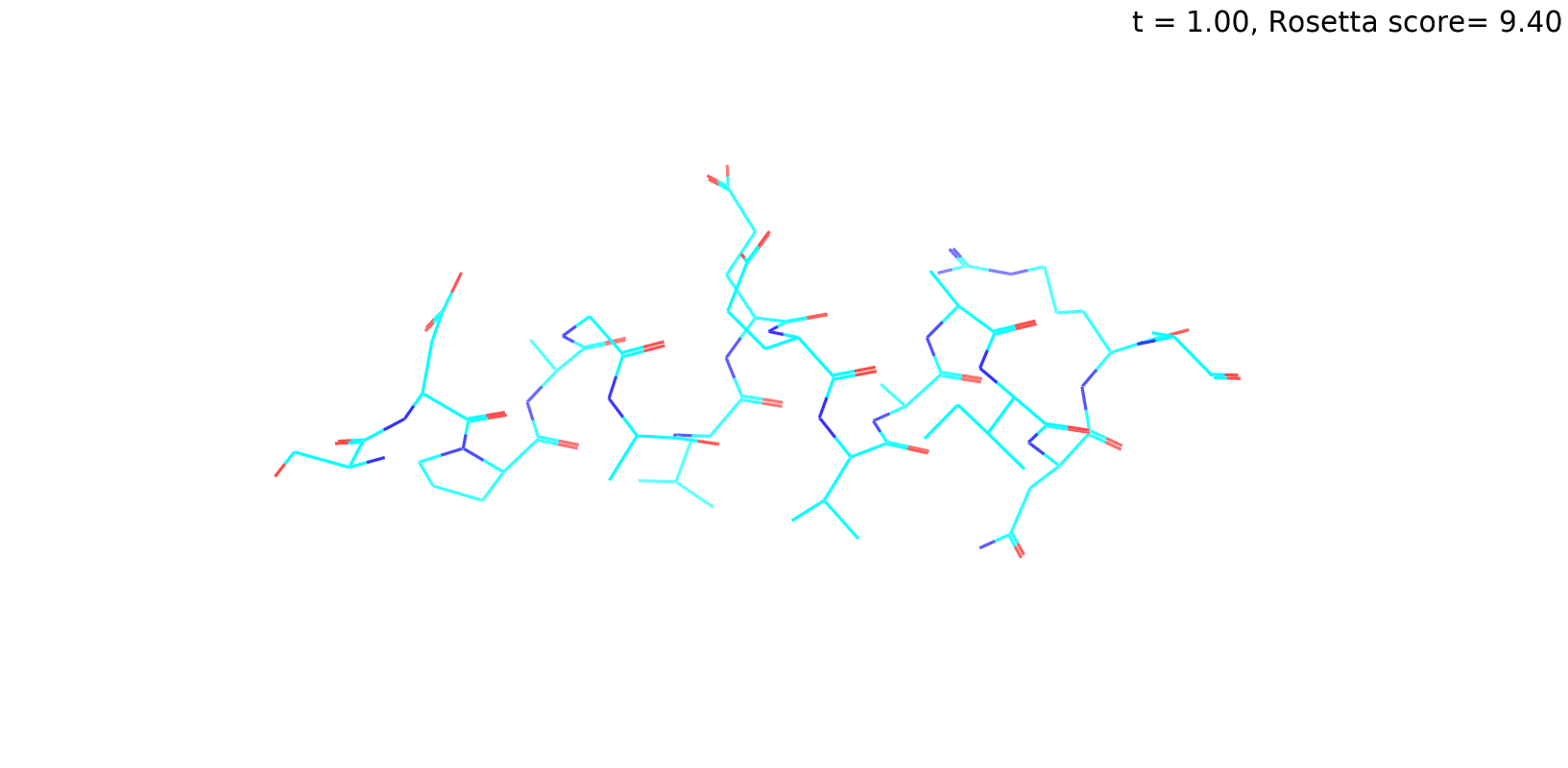
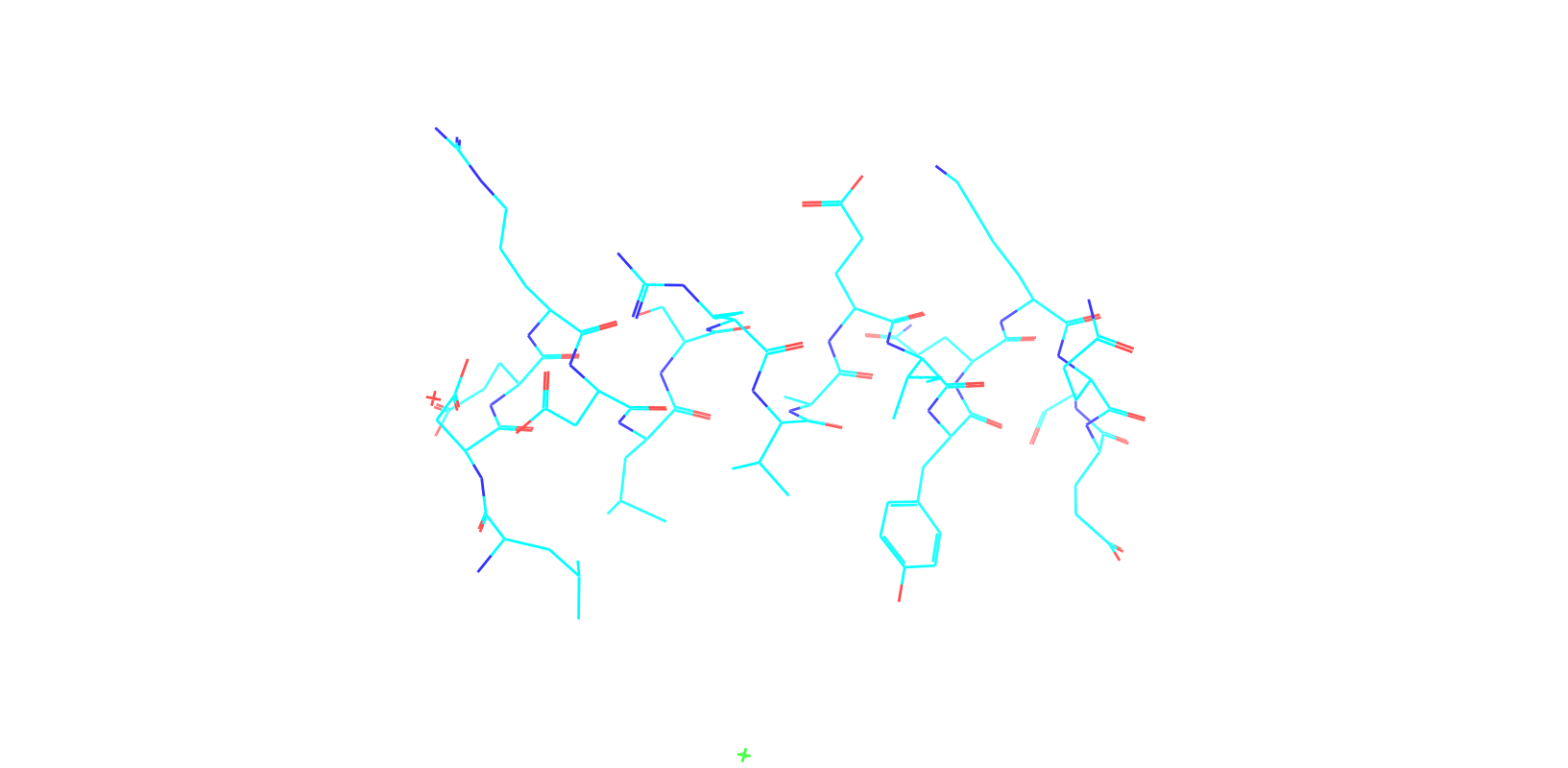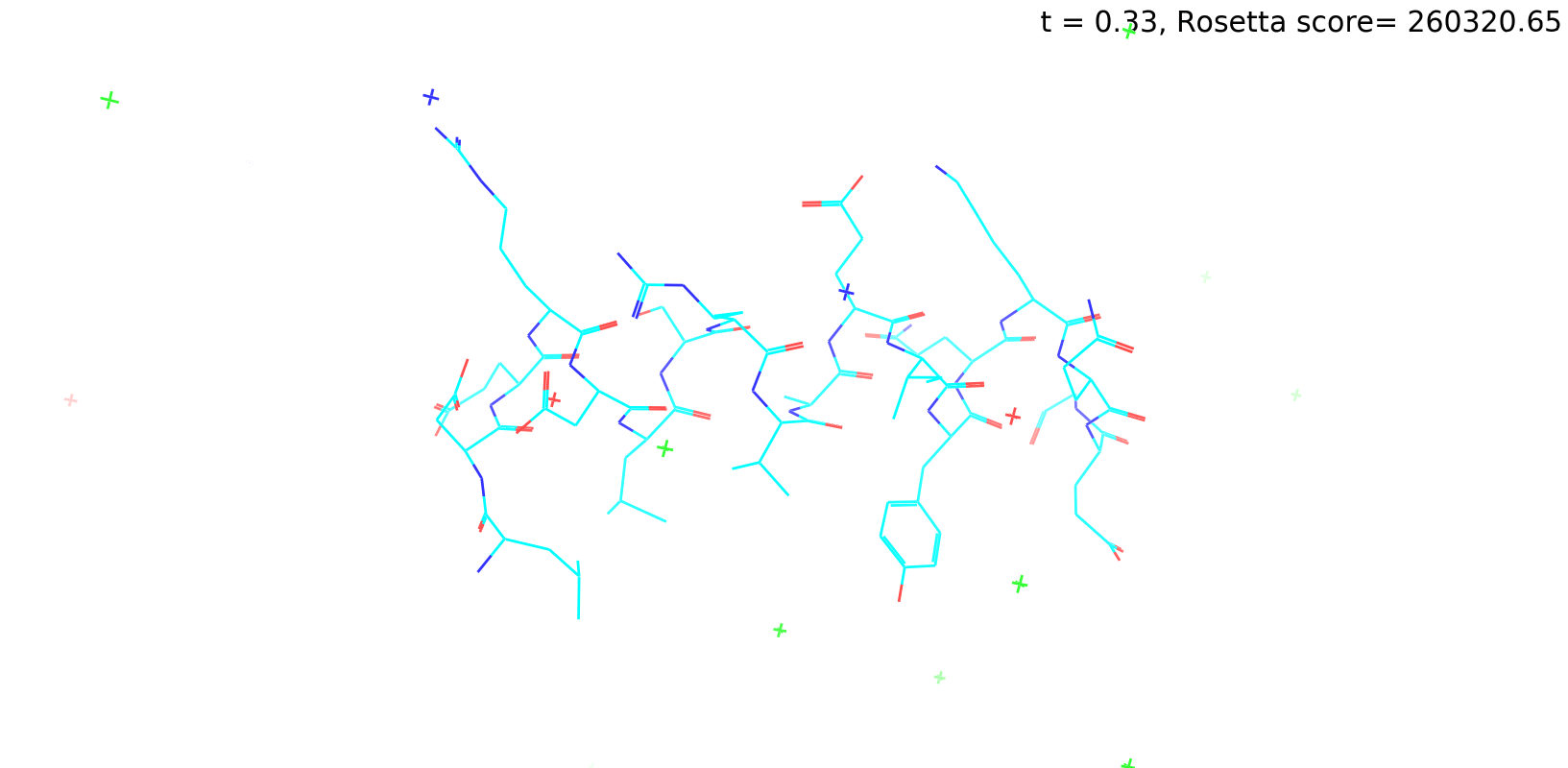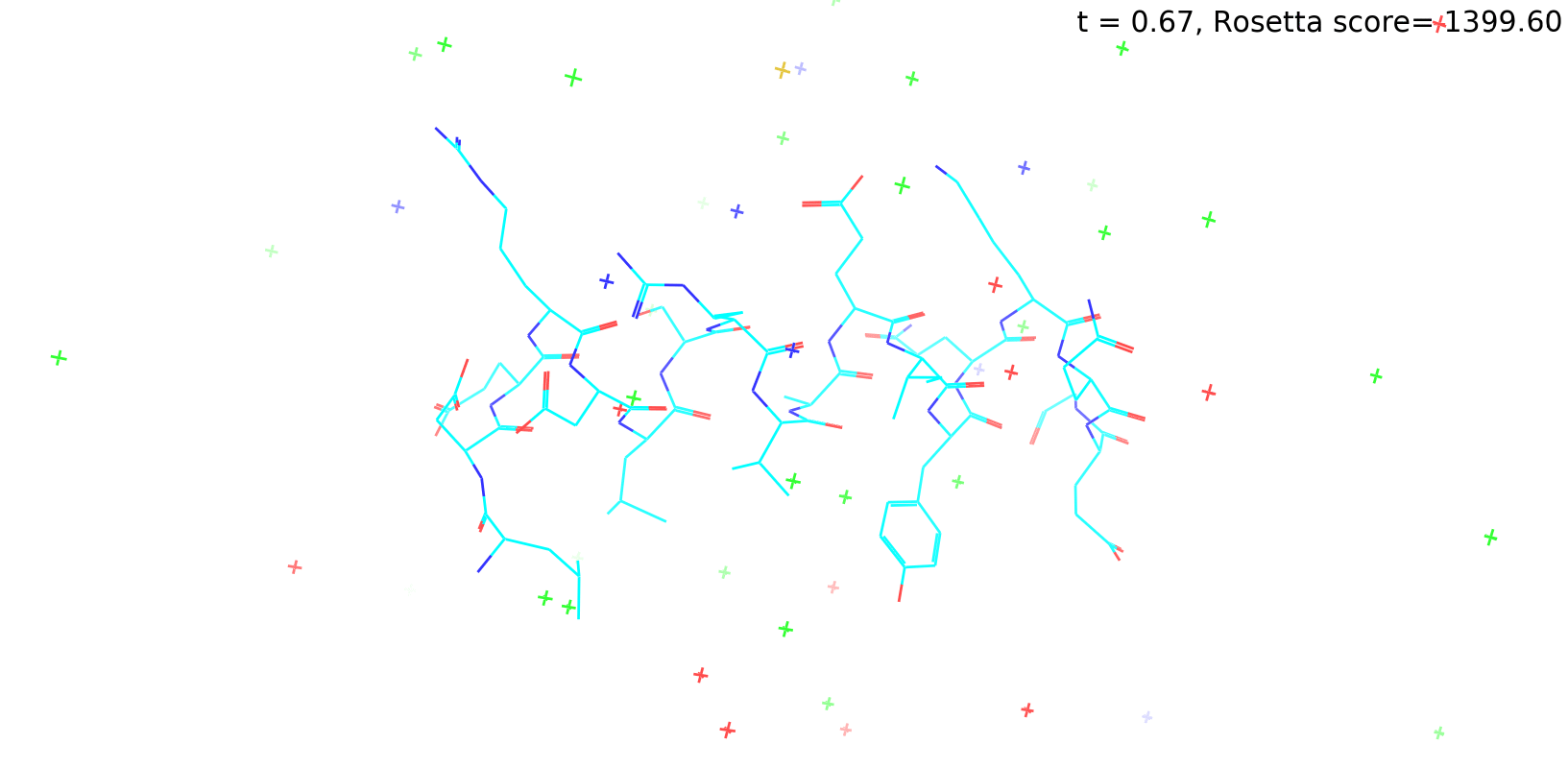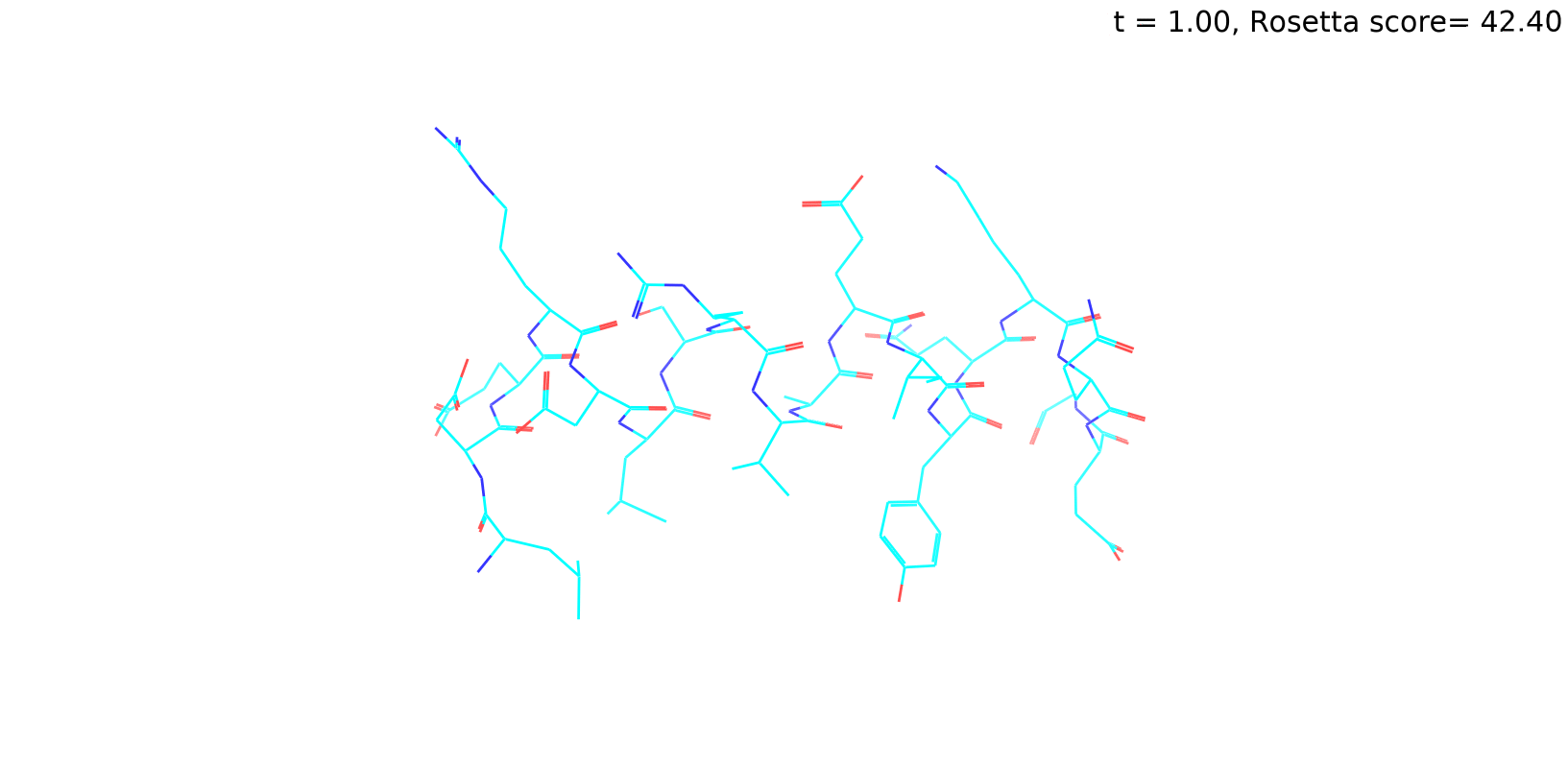
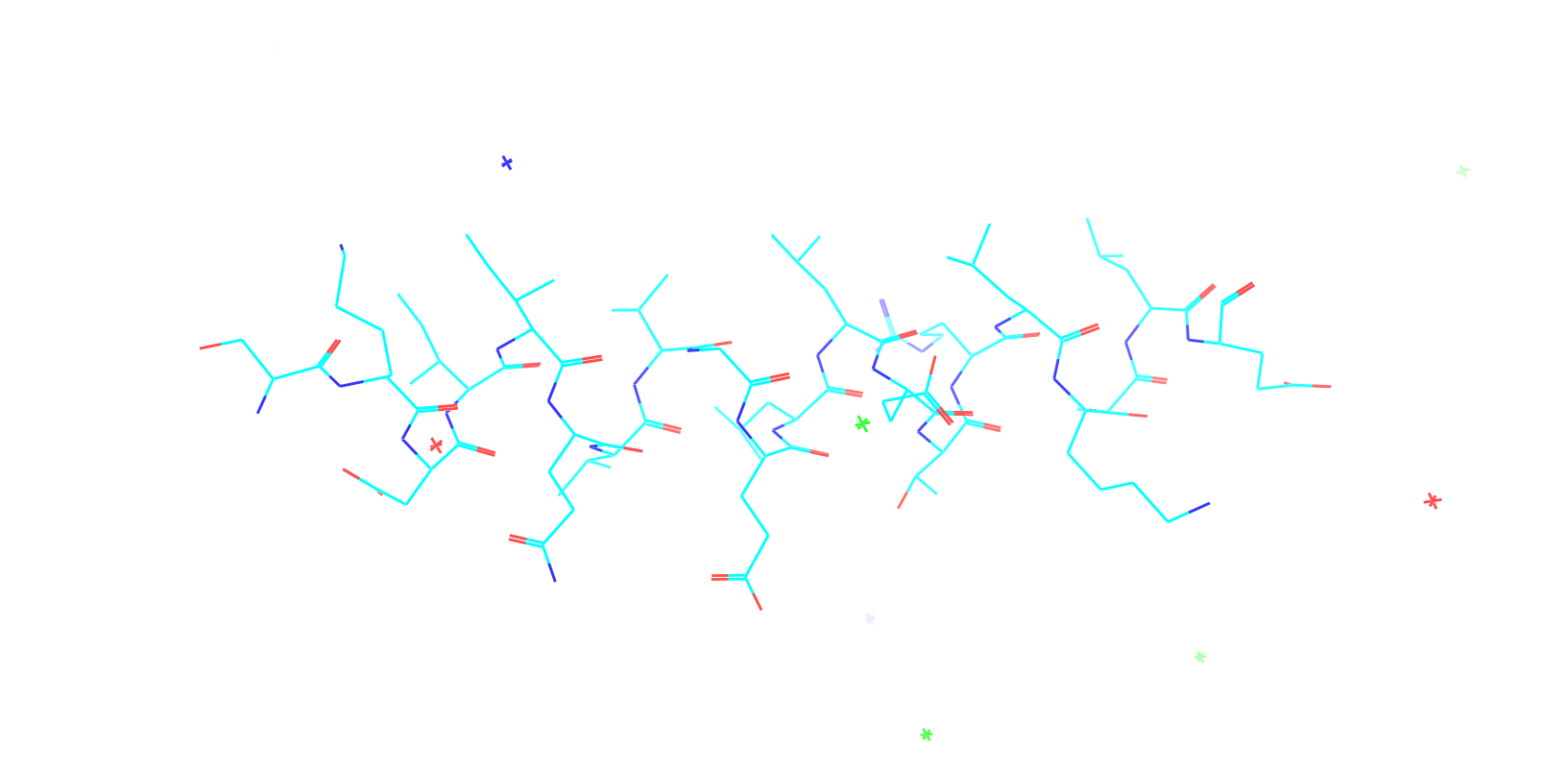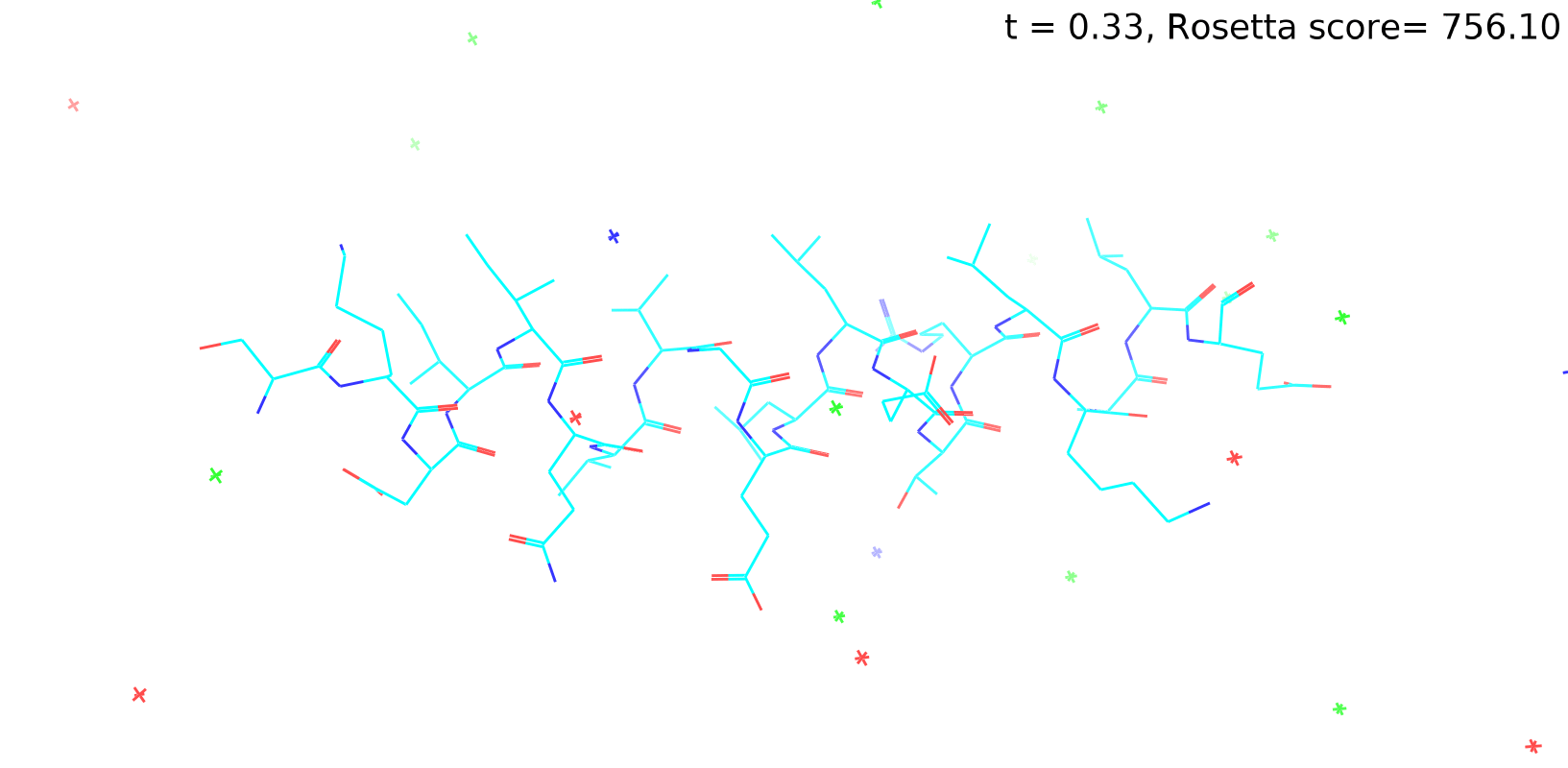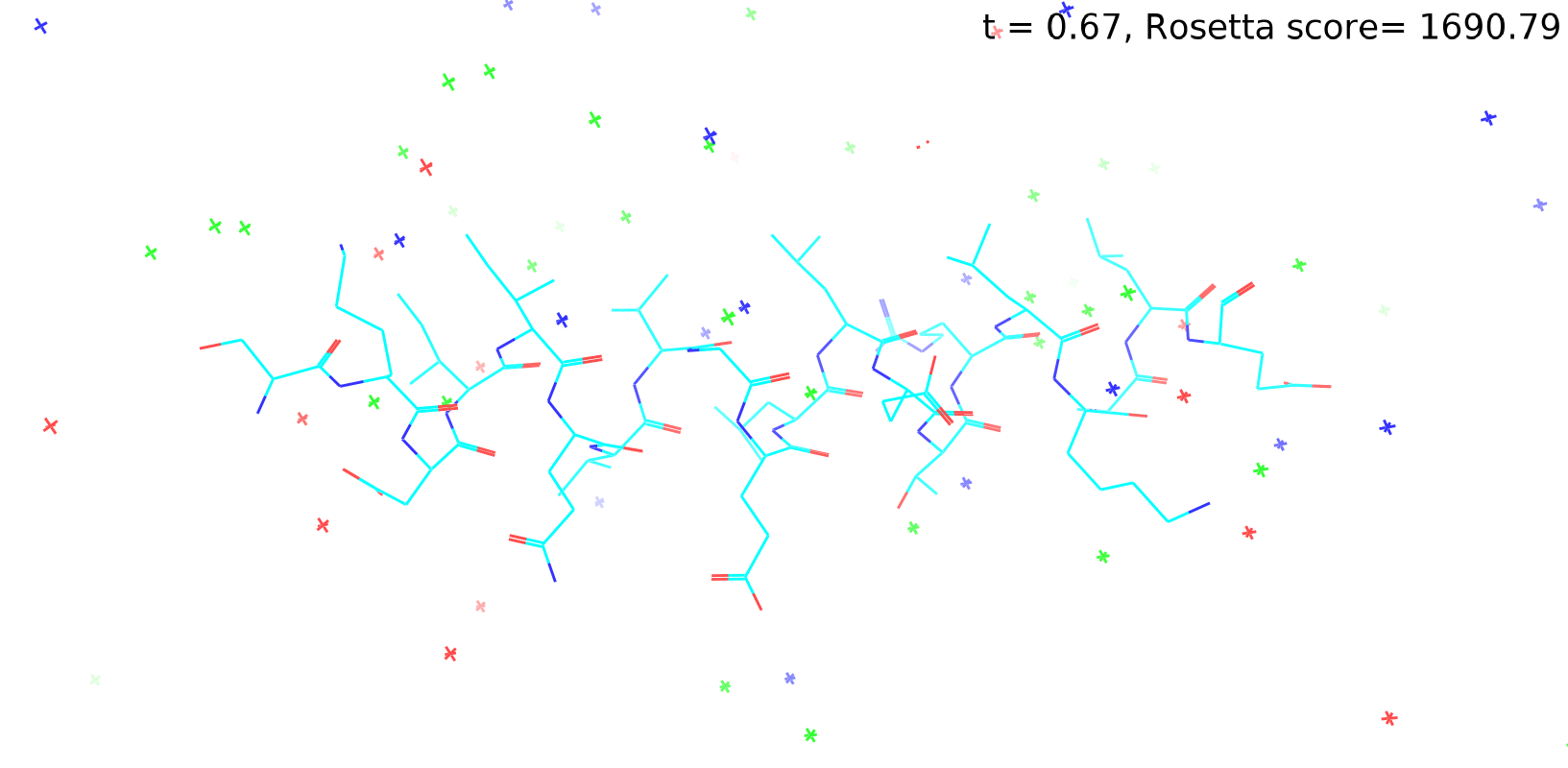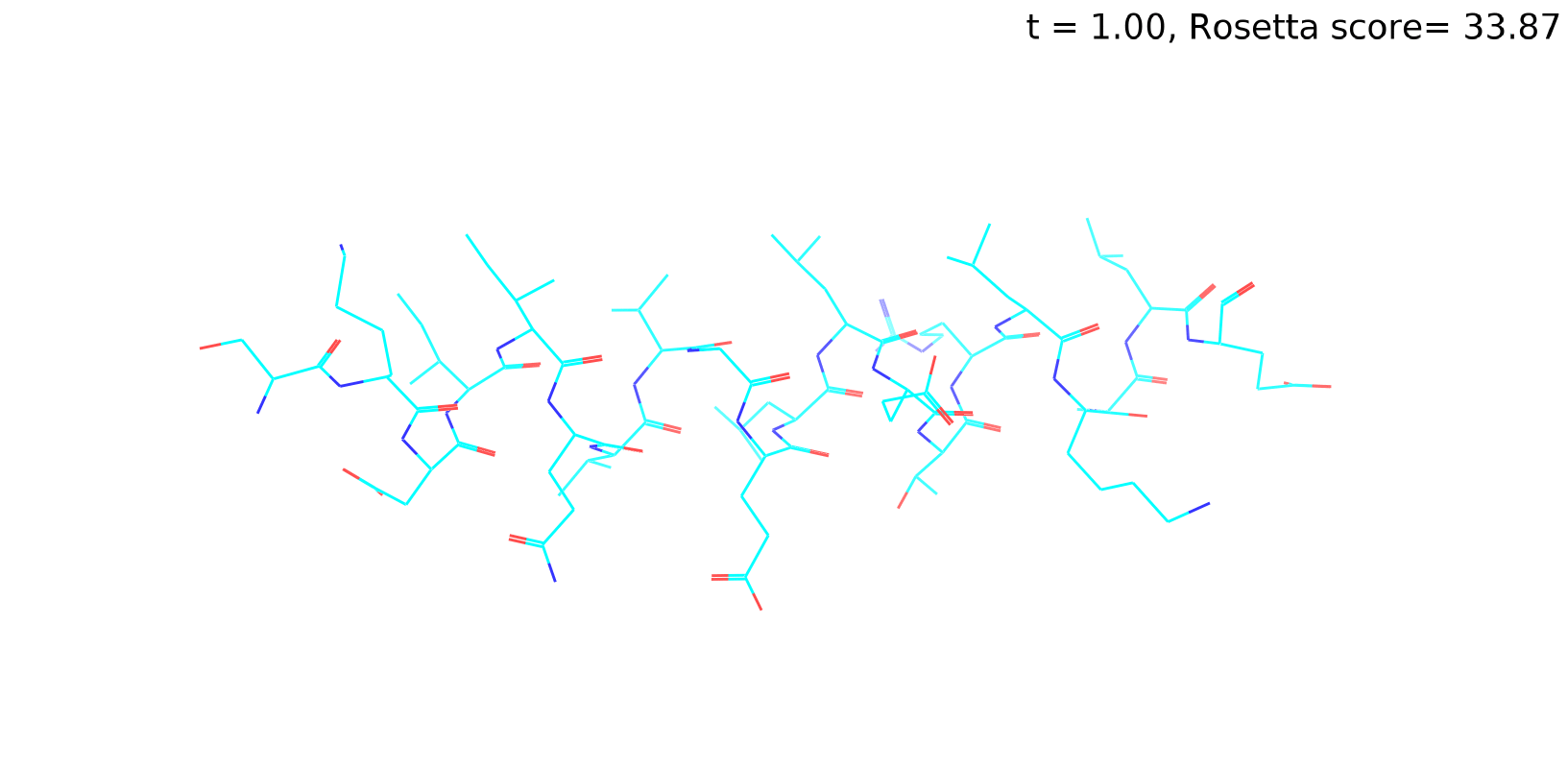
A pratice talks cover three my recent works (helixDiff, HelixFlow, and ABGM) on May 2024 and a visulization for new antibody project using flow-matching.
Resume
Xuezhi Xie
PhD student, Department of Computer Science, University of Toronto
Education
- Ph.D in Computer Science, University of Toronto, 2020 - 2025 (expected)
- M.S. in Computer Science, specialization in Artificial Intelligence, Western University, 2018 - 2020
- B.S. in Biology, Minor in Computer science, University of Waterloo, 2012 - 2016
Skills
- Languages: Java, Python, C, C#, C++, JavaScript
- Software: Eclipse, Jupyter, Visual Studio/Code
- Database: SQL, MongoDB
- Frameworks: Keras, Pytorch, Tensorflow, PySpark, Hadoop MapReduce
- System: Linux, Windows
- Version Control: Git/GitHub
- Machine learning: Proficient in deep generative models (diffusion models, gan, VAE), CNN, LSTM, logistic regression, support vector machine, decision tree, random forest, GBDT, naive bayes, k-means, PCA, collaborative filtering.
- Computer vision: Skilled in image processing (OpenCV), object detection, object segmentation, mapping and localization, and SLAM.
- Natural language processing: Skilled in text classification, sentiment analysis and speech recognition
Work experience
- Research assistant (machine learning) - University of Toronto, Toronto, On, Canada, 2020 - present
- Developed different deep generative model for target-pecific drug design, including score-based diffusion model, GAN, and VAE.
- Developed and implemented various searches and inpainting methods to optimize the synthetic data with optimized pharmetutical properties.
- Supervisor: Professor Philip M. Kim
- AI developer & Research assistant - Kaizhong’s lab, Western University, London, On, Canada, Sep.2018 - Apr. 2020
- Collected peptide data from online databases (IEDB, IPD-MHC). Preprocessed, and analyzed data by using PySpark
- Implemented and compared decision tree- and neural network-based models for predicting peptide binding.
- Designed and developed a novel CNN-LSTM model to solve a mhc-ligand binding classification task. Achieved state of the art performance (AUC : 92.3%).
- Published the work as first author in IEEE-BIBM (2019) and IJDMB(2020).
- Supervisor: Professor Kaizhong Zhang
Projects
- Modified AlphaFold 3 model for macrocyclic peptides design (Feb.2024 - Now)
- Developed CyclicBoltz1, an AlphaFold3-based model that extends cyclic offset encoding to predict cyclic peptide structures with non-canonical amino acids. Achieved superior accuracy over all current cyclic prediction models. Link for my paper
- Flow-matching model for antibody design (Jan.2024 - Dec.2024)
- Designed flow-matching model with equivariance for full-atom antibody design, and developed an active inpainting model for conditional design based on antigen.
- Score-based diffusion model for peptide drug design (Jan.2023 - Dec.2023)
- Designed score-based diffusion model for full-atom peptide design, and developed an active inpainting model for conditional design based on hotspots residues to facilitate early drug discovery. Link for my paper
- Invited to design the cover for ACS Central Science journal. Link for cover
- Diffusion model for antigen-specific antibody design (Jan.2022 - Dec.2023)
- Developed a score-based diffusion model named AntibodySGM for antigen specefic antibody design and achived state-of-art performance compared with current deep learning models.
- Developed and implemented a novel CDR-inpainting module for antigen-specific antibody optimization. Published the work as first author in ICML Workshop (ICML computational biology) 2023. Link for my paper
- Create an online visulization tools for peptide and protien. Generated antiboy data also included. Link for my visulization tool
- Deep generative model with constraints for D-peptide drug design (Jan.2019 - Jan.2023)
- Developed a deep generative model using GAN for novel dextrorotary helical conformations to facilitate the peptide drug design.
- Developed and implemented various latent search methods for the synthetic data with optimized pharmetutical properties. Published the work as first author in both Bioinformatics (2023) Link for my paper and NeuIPS Workshop (MLSB) 2021. Link for my talk
- Cell detection application for pathological image analysis (Jan.2019 - April.2020)
- Designed and developed customized object detection algorithms to detect brain cells and mitosis cells on pathology segmented images and whole slide images by using deep Convolutional Neural Network.
- Designed and developed Ki-67 cell segmentation and detection algorithm based on cellpose and image classification.
- Recommendation system for prediction of user purchase behavior (Jan. 2019 - Mar.2019)
- Classified data as train sets, validation sets and test sets. Constructed features using Pandas. Dealt with positive and negative sample imbalances using k-means and subsample.
- Used the Gradient Boosting Decision Tree to predict user purchase behavior through model training, parameter tuning and performance evaluation using F1-Score. Ranked 4th in 135 submission teams (Kaggle leaderboard link)
- Data Mining for Twitter Unstructured Data (Sep.2018 - Dec.2018)
- Mining interesting information from twitter tweets (JSON). Operated complex and unstructured data using MongoDB. Used MapReduce to process and summarize information. Conducted ElasticSearch. Visualized the information using Kibana.
Publications (First Author only)
jourals
Xie et al. “CyclicBoltz1, fast and accurately predicting structures of cyclic peptides and complexes containing non-canonical amino acids using AlphaFold 3 Framework”Link for my paper, bioRxiv, 2025.
Xie et al. “Antibody-SGM, a Score-Based Generative Model for Antibody Heavy-Chain Design”(link), Journal of Chemical Information and Modeling, 2024. Link for my paper
Xie et al. “HelixDiff, a Score-Based Diffusion Model for Generating All-Atom α-Helical Structures”(link), ACS Central Science (IF 18.2), 2024. Link for my paper
Xie et al. “HelixGAN a deep-learning methodology for conditional de novo design of α-helix structures”(link), Bioinformatics 2023.Link for my paper
Xie et al. “MHCherryPan, a novel pan-specific model for binding affinity prediction of class I HLA-peptide” (link), Int. J. Data Mining and Bioinformatics, Vol. 24, No. 3, 2020. Link for my paper
Conferences & workshops
- Xie et al. “HelixFlow, SE(3)–equivariant Full-atom Design of Peptides With Flow-matching Models”, Machine Learning for Structural Biology (MLSB) Workshop at NeurIPS 2024 .Link for my paper Link for my talk.
Xie et al. “HelixDiff: Conditional Full-atom Design of Peptides With Diffusion Models”(link), Machine Learning for Structural Biology (MLSB) Workshop at NeurIPS 2023 Link for my paper
Xie et al. “Antibody-SGM: Antigen-Specific Joint Design of Antibody Sequence and Structure using Diffusion Models”, Computational Biology Workshop at ICML 2023 Link for my paper
Xie et al. “HelixGAN: A bidirectional Generative Adversarial Network with search in latent space for generation under constraints”(link), Machine Learning for Structural Biology (MLSB) Workshop at NeurIPS 2021. Link for my paper Link for my talk
- Xie, et al. “MHCherryPan, a novel model to predict the binding affinity of pan-specific class I HLA-peptide” (link), IEEE International Conference on Bioinformatics and Biomedicine (IEEE - BIBM) 2019. Link for my paper